Similar to the last heatmap post I just revised, here I wrapped up the function and showing COVID-19 data just downloaded from JHU CSSE Github page. The function can take a given dataset and plot designated variable.
Code hosted on my Github repo.
US
# major function, can download from Github repo Blogdown/hugo-xmag/Code
source(here::here("Code/COVID_make_map.R"))
# US
dt_JUH_US <- get.JHU.us.state()
make_heatmap(data = dt_JUH_US,
geo_data = get_state_name(),
state_var = "Province_State", fill_var = "Mortality_Rate", label_var = "abb")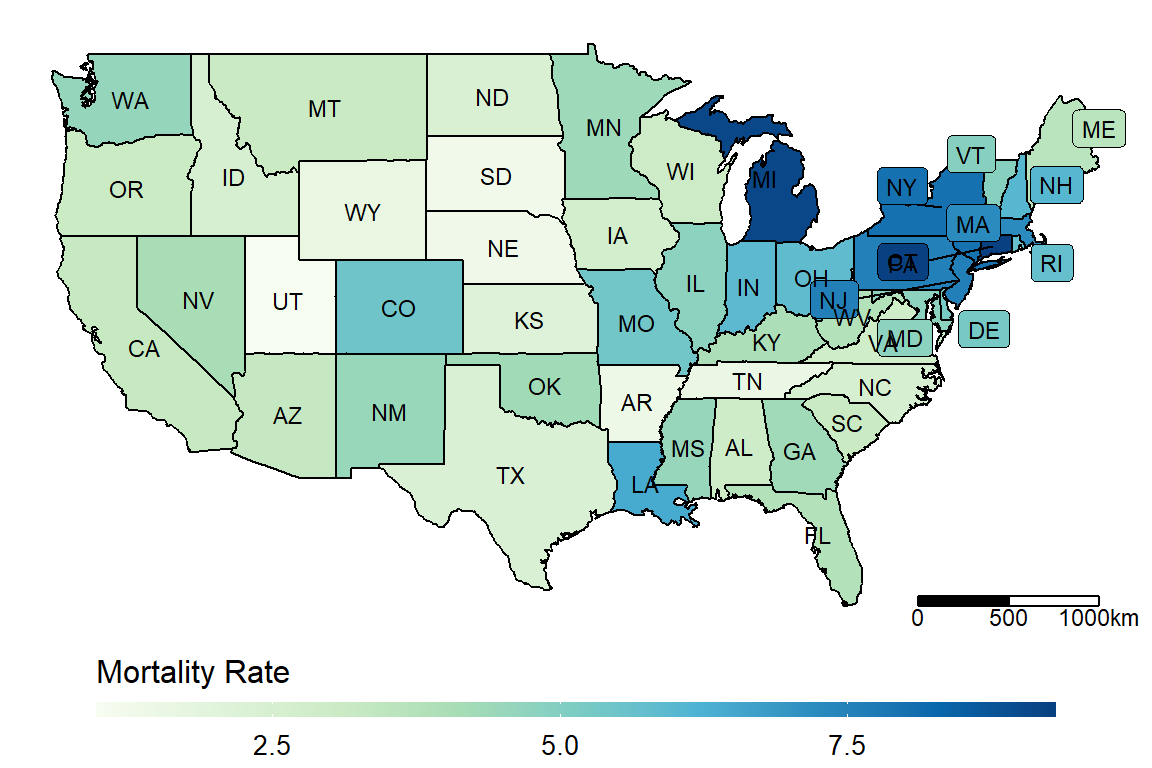
us_maps <- lapply(c("Mortality_Rate", "Incident_Rate", "Testing_Rate", "Hospitalization_Rate"),
make_heatmap,
data = dt_JUH_US,
geo_data = get_state_name(),
state_var = "Province_State", label_var = "abb")plot_grid <- gridExtra::grid.arrange(grobs = (us_maps), ncol = 2)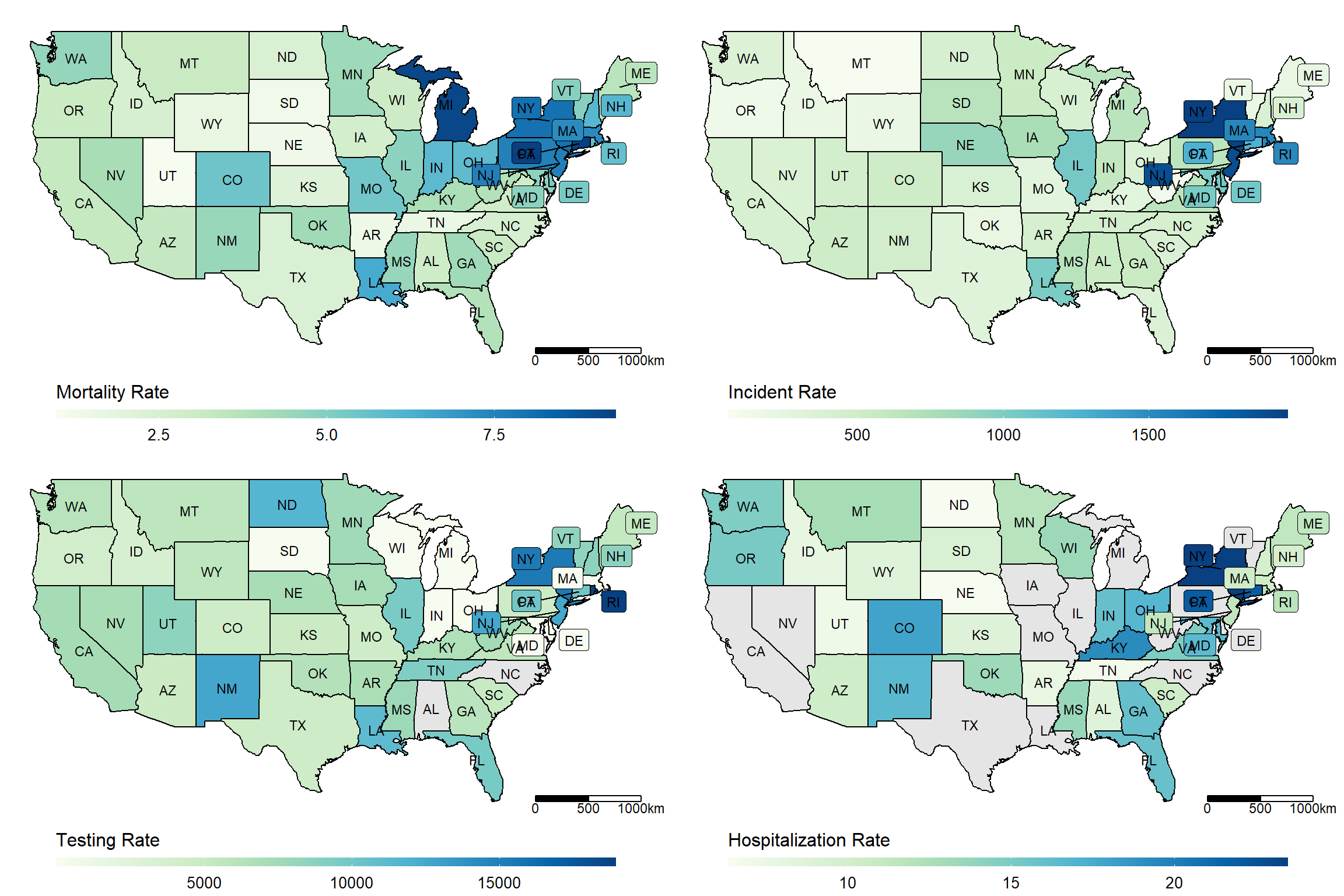
Compared to
China
# China (incl. Taiwan)
dt_JUH <- get.JHU.daily()
dt_CN <- dt_JUH[Country_Region %in% c("China", "Taiwan*"),]
dt_CN[Country_Region=="Taiwan*", Province_State:= "Taiwan"]
dt_china_map <- readRDS(here::here("../Data/dt_china.rds"))
setnames(dt_china_map, "province_EN", "state")
unique(dt_CN$Province_State[!dt_CN$Province_State%in%dt_china_map$state])## [1] "Hong Kong" "Macau" "Unknown"make_heatmap(data = dt_CN,
geo_data = dt_china_map,
state_var = "Province_State", fill_var = "Case-Fatality_Ratio")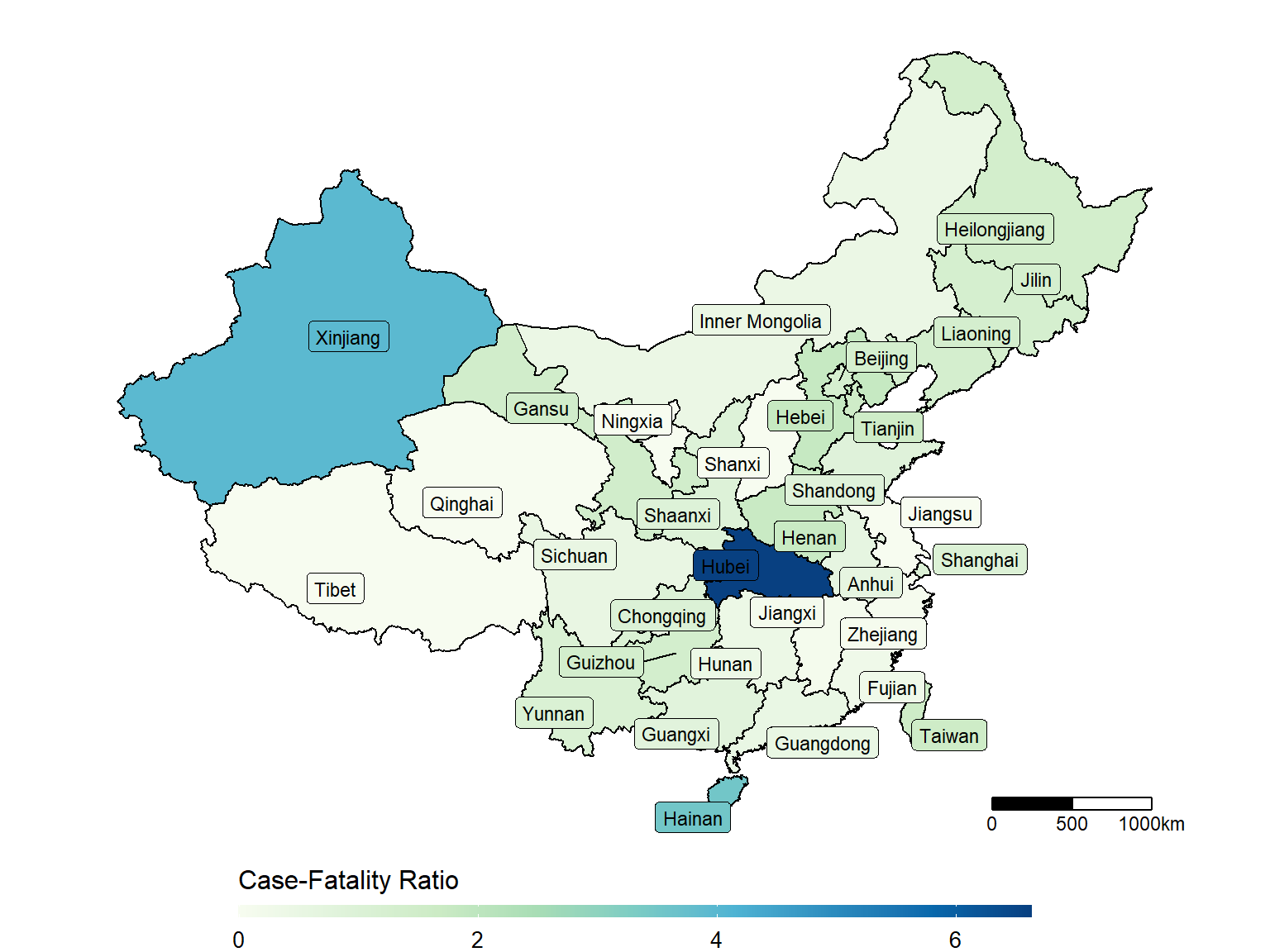
make_heatmap(data = dt_CN,
geo_data = dt_china_map,
state_var = "Province_State", fill_var = "Incidence_Rate")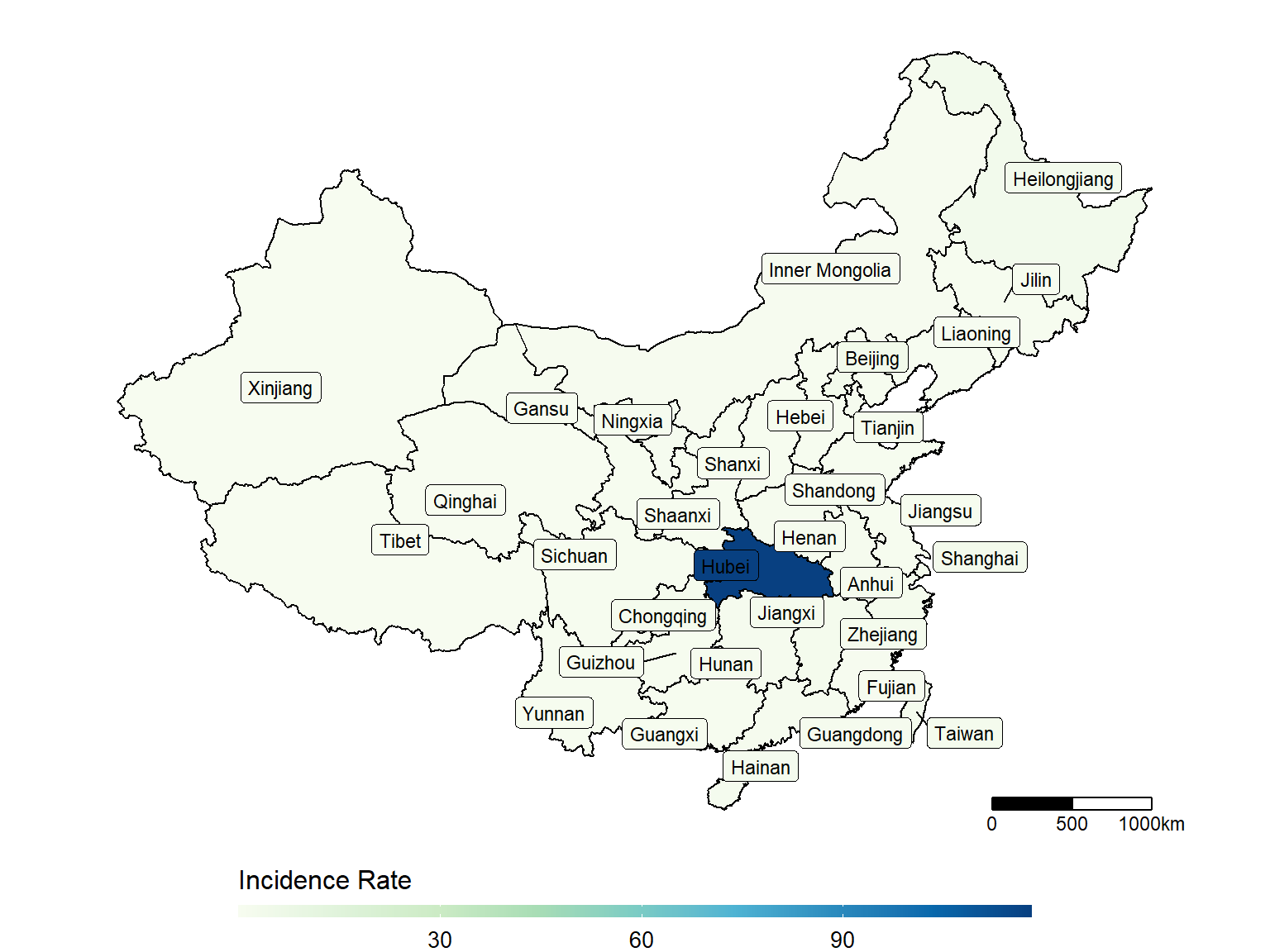
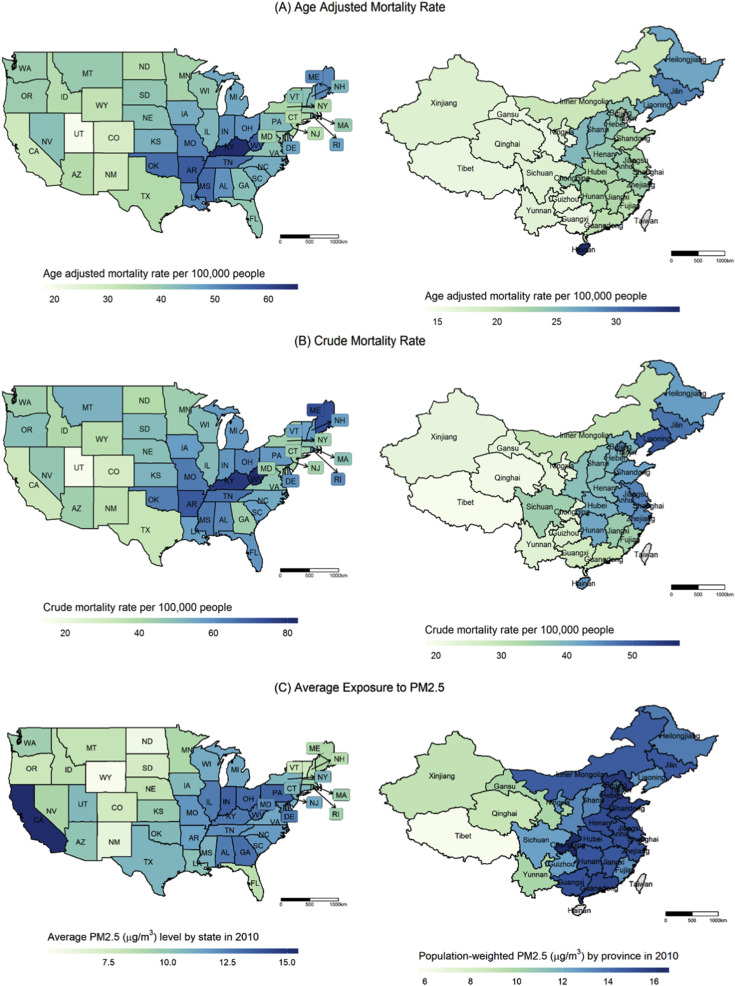
Compared to lung cancer mortality
Age-adjusted mortality rate (A) and crude mortality rate (B) in the US by state (in 2015) and in China by province (in 2008), from my paper “Epidemiology of lung cancer and lung cancer screening programs in China and the United States”, Cancer Letters 2020-01