A wrapped function to make summary plot from model object and predictors
Source:R/SHAP_funcs.R
shap.plot.summary.wrap1.Rdshap.plot.summary.wrap1 wraps up function shap.prep and
shap.plot.summary
Arguments
- model
the model
- X
the dataset of predictors used for calculating SHAP
- top_n
how many predictors you want to show in the plot (ranked)
- dilute
being numeric or logical (TRUE/FALSE), it aims to help make the test plot for large amount of data faster. If dilute = 5 will plot 1/5 of the data. If dilute = TRUE or a number, will plot at most half points per feature, so the plotting won't be too slow. If you put dilute too high, at least 10 points per feature would be kept. If the dataset is too small after dilution, will just plot all the data
Examples
# Example: Basic workflow for SHAP summary plot
# Note: For xgboost 3.x, use xgb.DMatrix + xgb.train, and convert factor labels to numeric
data("iris")
X1 = as.matrix(iris[,1:4])
y1 = as.numeric(iris[[5]]) - 1 # Convert factor to numeric
dtrain = xgboost::xgb.DMatrix(data = X1, label = y1)
params = list(learning_rate = 1, min_split_loss = 0, reg_lambda = 0,
objective = 'reg:squarederror', nthread = 1)
mod1 = xgboost::xgb.train(params = params, data = dtrain,
nrounds = 1, verbose = 0)
# Get SHAP values and feature importance
shap_values <- shap.values(xgb_model = mod1, X_train = X1)
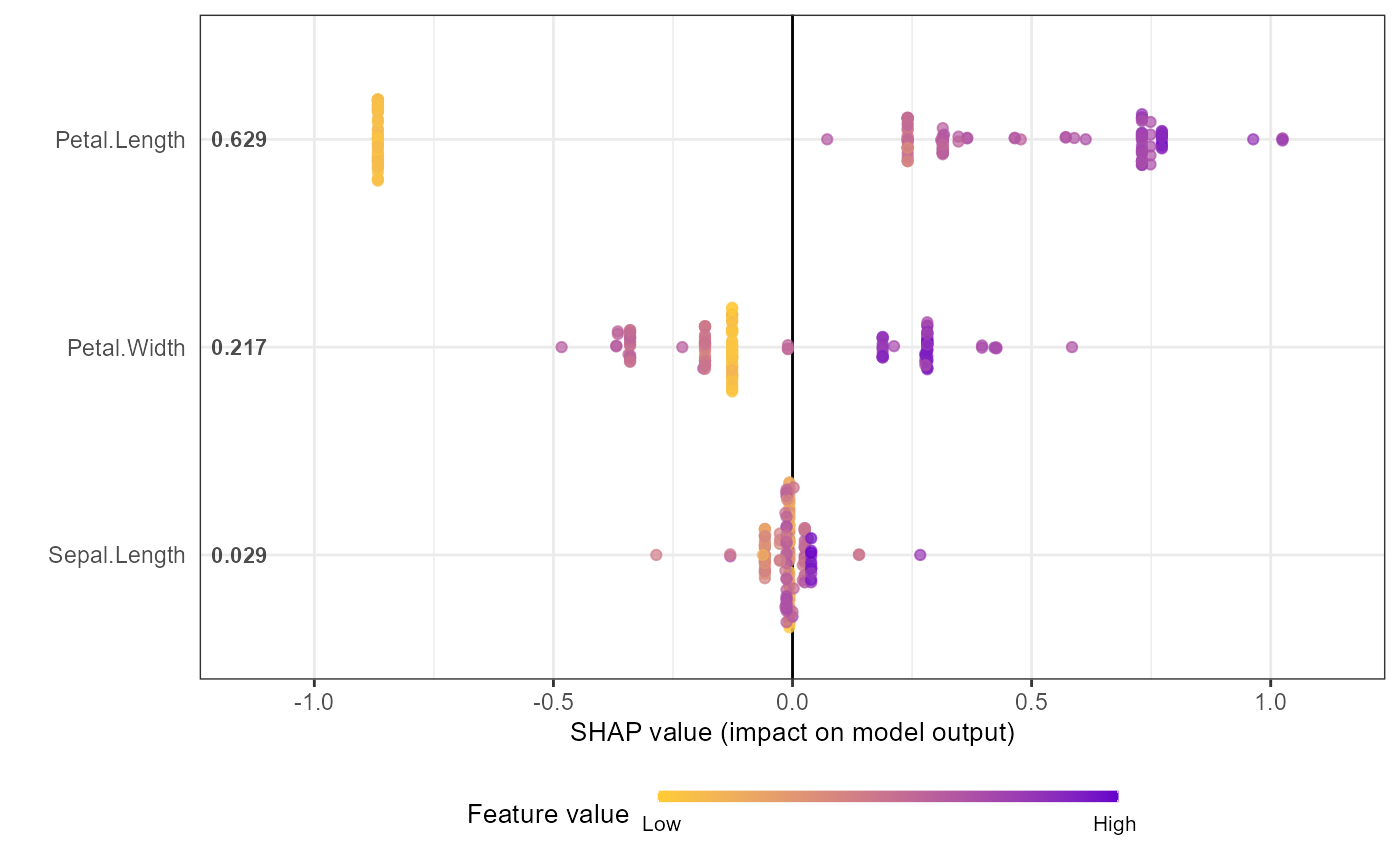
shap_values$mean_shap_score # Ranked features by mean|SHAP|
#> Petal.Length Petal.Width Sepal.Length Sepal.Width
#> 0.6307042 0.2135736 0.0300757 0.0000000
shap_values_iris <- shap_values$shap_score
# Prepare long-format data for plotting
shap_long_iris <- shap.prep(xgb_model = mod1, X_train = X1)
# Alternative: use pre-computed SHAP values
shap_long_iris <- shap.prep(shap_contrib = shap_values_iris, X_train = X1)
# SHAP summary plot
shap.plot.summary(shap_long_iris, scientific = TRUE)
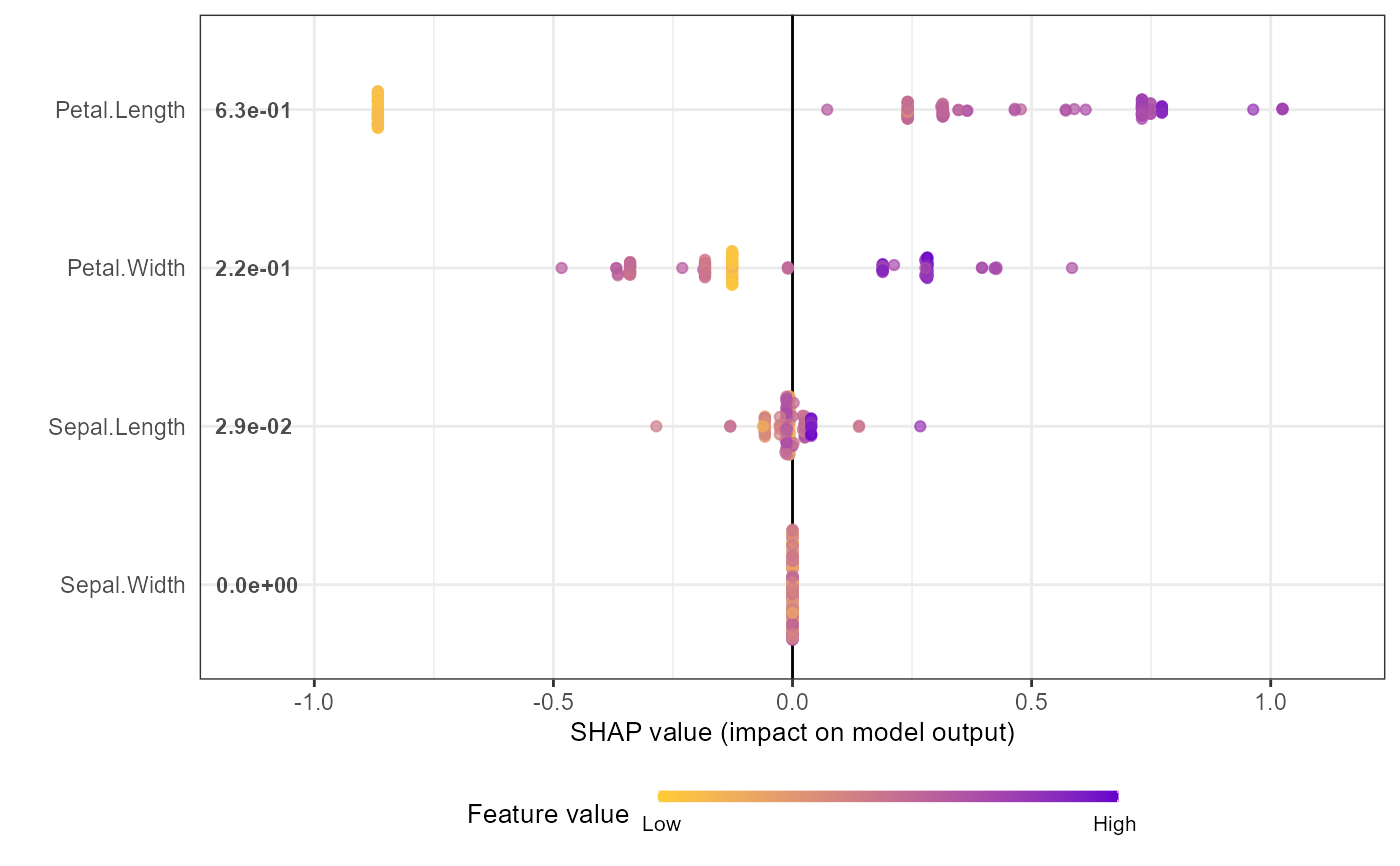 shap.plot.summary(shap_long_iris, x_bound = 1.5, dilute = 10)
shap.plot.summary(shap_long_iris, x_bound = 1.5, dilute = 10)
 # Alternative options:
# Option 1: directly from xgboost model
shap.plot.summary.wrap1(mod1, X = as.matrix(iris[,1:4]), top_n = 3)
# Alternative options:
# Option 1: directly from xgboost model
shap.plot.summary.wrap1(mod1, X = as.matrix(iris[,1:4]), top_n = 3)
 # Option 2: from pre-computed SHAP values (useful for cross-validation)
shap.plot.summary.wrap2(shap_score = shap_values_iris, X = X1, top_n = 3)
# Option 2: from pre-computed SHAP values (useful for cross-validation)
shap.plot.summary.wrap2(shap_score = shap_values_iris, X = X1, top_n = 3)